Robetta is a protein structure prediction service that is continually evaluated through CAMEO
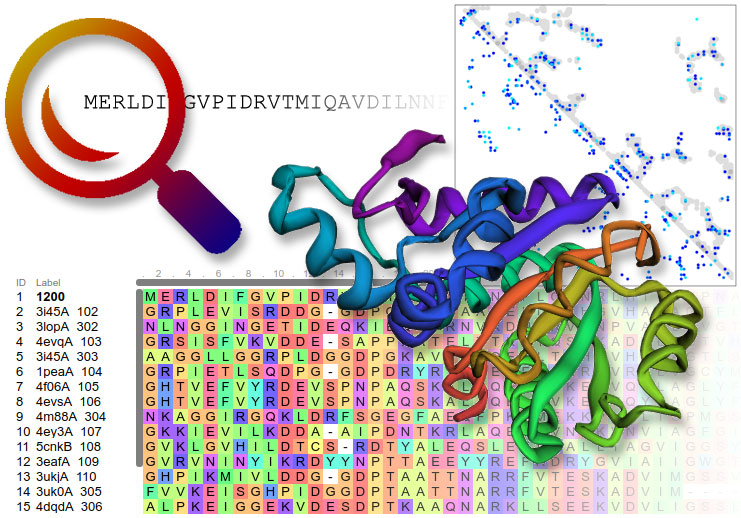
Features include relatively fast and accurate deep learning based method, RoseTTAFold, and an interactive submission interface that allows custom sequence alignments for homology modeling, constraints, local fragments, and more. It can model multi-chain complexes using RoseTTAFold (user must provide paired MSA) or comparative modeling (CM) and provides the option for large scale sampling. The CM method uses the PDB100 template database, a co-evolution based model database (MDB), and also provides the option for custom templates. Computing resources are provided by the Baker lab and by volunteers from the distributed computing project Rosetta@home. You can help this service by joining Rosetta@home.
For more information please visit our Frequently Asked Questions.
PDB templates last updated June 14 2024
Jobs queued: 29 active: 612
Users: 74742 Countries: 233 New users last week: 152 New jobs last week: 101 New final models last week: 364
Last updated February 26 2026 13:01
Baker Lab | Rosetta@home | Contact | Terms of Service
©2026 University of Washington